Subacute Necrotizing Encephalomyelopathy and MT-CO1
MT-CO1 Motifs
The MT-CO1 gene (human) was used to determine whether and which motifs there were within the gene. The file containing the sequence is below.
| mt-co1_gene_sequence.docx |
Restriction Endonuclease Map
The restriction endonuclease sites available in the MT-CO1 gene sequence are listed below in Bold and Underlined. The gene sequence in which it cuts is following in italics, with the nucleotide count location in (parenthesis) [3,4].
AccIII T^CCGGA (785, 1301)
AclI AA^CGTT (165)
AhlI A^CTAGT (244)
Aor13HI T^CCGGA (785, 1301)
AseI AT^TAAT (1400)
AspA2I C^CTAGG (85, 847, 1024)
AsuII TT^CGAA (1433, 1515)
AvrII C^CTAGG (85, 847, 1024)
BcuI A^CTAGT (244)
BglI GCCNNNN^NGGC 2 (132, 1062)
BlnI C^CTAGG (85, 847, 1024)
Bpu14I TT^CGAA (1433, 1515)
BseAI T^CCGGA (785, 1301)
Bsp119I TT^CGAA (1433, 1515)
Bsp13I T^CCGGA (785, 1301)
BspEI T^CCGGA (785, 1301)
BspHI T^CATGA (1410)
BspT104I TT^CGAA (1433, 1515)
BssNAI GTA^TAC (1528)
Bst1107I GTA^TAC (1528)
BstBI TT^CGAA 2(433, 1515)
BstEII G^GTNACC (716)
BstPI G^GTNACC (716)
BstXI CCANNNNN^NTGG (1474)
BstZ17I GTA^TAC (1528)
CciI T^CATGA (1410)
Csp45I TT^CGAA (1433, 1515)
Eco32I GAT^ATC (834)
AclI AA^CGTT (165)
AhlI A^CTAGT (244)
Aor13HI T^CCGGA (785, 1301)
AseI AT^TAAT (1400)
AspA2I C^CTAGG (85, 847, 1024)
AsuII TT^CGAA (1433, 1515)
AvrII C^CTAGG (85, 847, 1024)
BcuI A^CTAGT (244)
BglI GCCNNNN^NGGC 2 (132, 1062)
BlnI C^CTAGG (85, 847, 1024)
Bpu14I TT^CGAA (1433, 1515)
BseAI T^CCGGA (785, 1301)
Bsp119I TT^CGAA (1433, 1515)
Bsp13I T^CCGGA (785, 1301)
BspEI T^CCGGA (785, 1301)
BspHI T^CATGA (1410)
BspT104I TT^CGAA (1433, 1515)
BssNAI GTA^TAC (1528)
Bst1107I GTA^TAC (1528)
BstBI TT^CGAA 2(433, 1515)
BstEII G^GTNACC (716)
BstPI G^GTNACC (716)
BstXI CCANNNNN^NTGG (1474)
BstZ17I GTA^TAC (1528)
CciI T^CATGA (1410)
Csp45I TT^CGAA (1433, 1515)
Eco32I GAT^ATC (834)
Eco91I G^GTNACC (716)
EcoO65I G^GTNACC (716)
EcoRV GAT^ATC (834)
EcoT22I ATGCA^T (187, 1339)
HindIII A^AGCTT (301)
Kpn2I T^CCGGA (785, 1301)
MfeI C^AATTG (837)
Mph1103I ATGCA^T (187, 1339)
MroI T^CCGGA 785, (1301)
MunI C^AATTG (837)
NsiI ATGCA^T (187, 1339)
NspV TT^CGAA (1433, 1515)
PagI T^CATGA (1410)
PshBI AT^TAAT (1400)
Psp1406I AA^CGTT (165)
PspEI G^GTNACC (716)
PstI CTGCA^G (1012)
RcaI T^CATGA (1410)
SfuI TT^CGAA (1433, 1515)
SgrAI CR^CCGGYG (947)
SpeI A^CTAGT (244)
SspI AAT^ATT (1398)
VspI AT^TAAT (1400)
XbaI T^CTAGA (1538)
XcmI CCANNNNN^NNNNTGG 1 (41)
XmaJI C^CTAGG (85, 847, 1024)
Zsp2I ATGCA^T (187, 1339)
EcoO65I G^GTNACC (716)
EcoRV GAT^ATC (834)
EcoT22I ATGCA^T (187, 1339)
HindIII A^AGCTT (301)
Kpn2I T^CCGGA (785, 1301)
MfeI C^AATTG (837)
Mph1103I ATGCA^T (187, 1339)
MroI T^CCGGA 785, (1301)
MunI C^AATTG (837)
NsiI ATGCA^T (187, 1339)
NspV TT^CGAA (1433, 1515)
PagI T^CATGA (1410)
PshBI AT^TAAT (1400)
Psp1406I AA^CGTT (165)
PspEI G^GTNACC (716)
PstI CTGCA^G (1012)
RcaI T^CATGA (1410)
SfuI TT^CGAA (1433, 1515)
SgrAI CR^CCGGYG (947)
SpeI A^CTAGT (244)
SspI AAT^ATT (1398)
VspI AT^TAAT (1400)
XbaI T^CTAGA (1538)
XcmI CCANNNNN^NNNNTGG 1 (41)
XmaJI C^CTAGG (85, 847, 1024)
Zsp2I ATGCA^T (187, 1339)
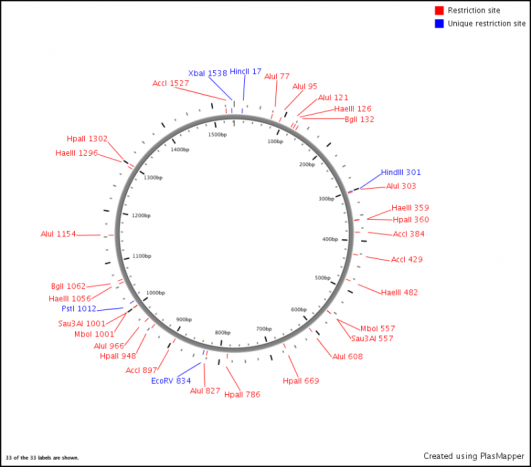
The restriction enzymes were detected and visually oriented on MT-CO1 using PlasMapper 2.0 [5]. This program is usually used with plasmids and has therefore oriented the gene circularly. This is anaccurate, because although MT-CO1 is found on a circular genome (Mitochondrial Genome) there is much more genes encoded both before and after the MT-CO1 gene. Despite the circular orientation, this is picture is hlpful in planning a restriction enzyme digest on the MT-CO1 gene.
Motif Search

GenomeNet Logo
Motif is a search engine able to identify motifs within amino acid and nucleotide sequences. The engine is maintained by GenomeNet, which harbors many other online biotechnology resources.
When a nucleotide sequence is analyzed, it is compared to the TRANSFAC database. TRANSFAC is a database based on eukaryotic cis-acting regulatory DNA elements and trans-acting factors . TRANSFAC links link to TFMATRIX database, with varying degrees of information regarding the motif. The cut off score is calculated using a formula based on the comparison of the motif sequence and the sequence in question [1].
When a nucleotide sequence is analyzed, it is compared to the TRANSFAC database. TRANSFAC is a database based on eukaryotic cis-acting regulatory DNA elements and trans-acting factors . TRANSFAC links link to TFMATRIX database, with varying degrees of information regarding the motif. The cut off score is calculated using a formula based on the comparison of the motif sequence and the sequence in question [1].
Motif Found (Symbol) and Transfac Link
Alcohol Dehydrogenase Gene Regulator 1 (ADR1)
M00048
GATA-Binding Factor 2 (GATA-2)
M00076
CdxA (CdxA)
M00101
arunt-factor AML-1 (AML-1)
M00271
activator of nitrogen-regulated genes (NIT2)
M00142
MZF1 (MZF1)
M00083
sex-determining region Y gene product (SYR)
M00148
c-Rel( c-Rel)
M00053
GATA-binding factor 1 (GATA-1)
M00075
stress-response element (STRE)
M00154
Lmo2 bound to Tal-1, E2A proteins, and GATA-1, half-site 2 (Lmo2)
M00278
GATA-binding factor 3 (GATA-3)
M00077
M00048
GATA-Binding Factor 2 (GATA-2)
M00076
CdxA (CdxA)
M00101
arunt-factor AML-1 (AML-1)
M00271
activator of nitrogen-regulated genes (NIT2)
M00142
MZF1 (MZF1)
M00083
sex-determining region Y gene product (SYR)
M00148
c-Rel( c-Rel)
M00053
GATA-binding factor 1 (GATA-1)
M00075
stress-response element (STRE)
M00154
Lmo2 bound to Tal-1, E2A proteins, and GATA-1, half-site 2 (Lmo2)
M00278
GATA-binding factor 3 (GATA-3)
M00077
Location (Cut off score)
685..680 (100)
667..662 (98)
1333..1328 (98)
1500..1495 (98)
271..266 (96)
945..936 (100)
1304..1295 (94)
1398..1404 (100)
1403..1397 (100)
65..59 (98)
212..206 (94)
37..32 (100)
157..152 (100)
1347..1342 (100)
1510..1505 (100)
590..595 (98)
471..476 (95)
948..941 (98)
291..297 (96)
1312..1303 (95)
945..936 (94)
475..482 (94)
269..277 (94)
595..587 (94)
667..662 (98)
1333..1328 (98)
1500..1495 (98)
271..266 (96)
945..936 (100)
1304..1295 (94)
1398..1404 (100)
1403..1397 (100)
65..59 (98)
212..206 (94)
37..32 (100)
157..152 (100)
1347..1342 (100)
1510..1505 (100)
590..595 (98)
471..476 (95)
948..941 (98)
291..297 (96)
1312..1303 (95)
945..936 (94)
475..482 (94)
269..277 (94)
595..587 (94)
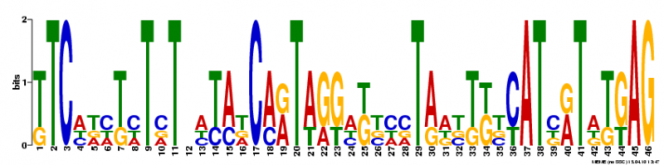
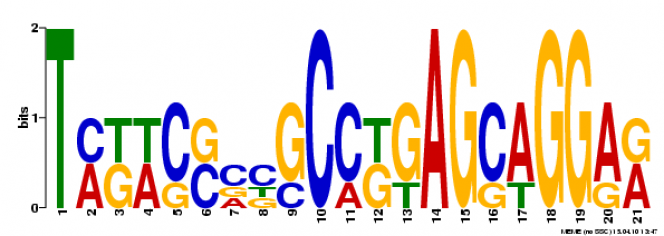
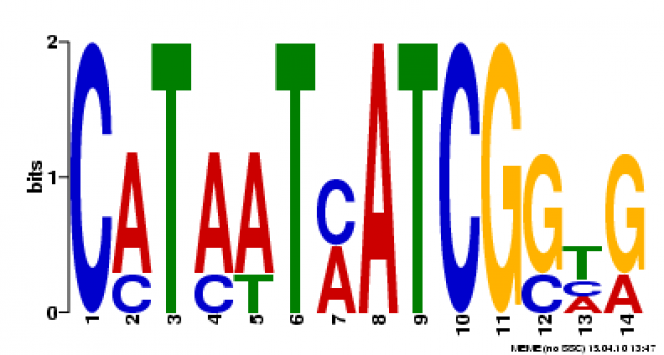
MEME Search

Multiple Em for Motif Elicitation (MEME) is a search engine that will analyze the gene sequence for similarities among and produce a description for each pattern it discovers [2]. It is maintained by an Opal Toolkit.
The motifs are not extrapolated into functional data of the DNA or protein, and therefore need to be searched in other motif databases in order to determine their significance. Therefore, motif comparison databases were used to determine whether these 3 motifs were present in biologically relevant molecules (TOMTOM, FIMO). To better understand the motifs found with FIMO, it was suggested to download the FIMO Sorftware (This was not done).
The motifs are not extrapolated into functional data of the DNA or protein, and therefore need to be searched in other motif databases in order to determine their significance. Therefore, motif comparison databases were used to determine whether these 3 motifs were present in biologically relevant molecules (TOMTOM, FIMO). To better understand the motifs found with FIMO, it was suggested to download the FIMO Sorftware (This was not done).
Motif 1:
This motif is found 4 times throughout the MT-CO1 gene sequence. The motifs is 46 base pairs long starting at 1128, 1374, 819, 1029 (E-values of 1.52e-17, 3.17e-17, 3.17e-17, 3.17e-17 respectively).
When using TOMTOM to search the JASPAR Core database, no comparative motifs were found under the q-value of 0.5 (default value). Using FIMO to search the Protein Data Bank, also yielded no results.
This motif is found 4 times throughout the MT-CO1 gene sequence. The motifs is 46 base pairs long starting at 1128, 1374, 819, 1029 (E-values of 1.52e-17, 3.17e-17, 3.17e-17, 3.17e-17 respectively).
When using TOMTOM to search the JASPAR Core database, no comparative motifs were found under the q-value of 0.5 (default value). Using FIMO to search the Protein Data Bank, also yielded no results.
Motif 2:
This motif is found 4 times throughout the MT-CO1 gene sequence. The motifs is 21 base pairs long starting at 658, 349, 61, 382 (E-values of 9.93e-12, 1.85e-11, 2.08e-10, 2.46e-09 respectively).
When using TOMTOM to search the JASPAR Core database, no comparative motifs were found under the q-value of 0.5 (default value). Using FIMO to search the Protein Data Bank, also yielded no results.
This motif is found 4 times throughout the MT-CO1 gene sequence. The motifs is 21 base pairs long starting at 658, 349, 61, 382 (E-values of 9.93e-12, 1.85e-11, 2.08e-10, 2.46e-09 respectively).
When using TOMTOM to search the JASPAR Core database, no comparative motifs were found under the q-value of 0.5 (default value). Using FIMO to search the Protein Data Bank, also yielded no results.
Motif 3: This motif is found 4 times throughout the MT-CO1 gene sequence. The motifs is 14 base pairs long starting at 251, 215, 1247, 926 (E-values of 3.11e-08, 7.47e-08, 9.24e-08, 2.96e-07 respectively).
When using TOMTOM to search the JASPAR Core database, one comparative motif was found. This molecule, ATHB-5, is found in Arabadosis thaliana. This is encouraging, because the Arabadopsis thaliana ortholog of the MT-CO1, was very similar to the human gene. It is possible the human ortholog was not within the database.
When using FIMO to search the Protein Data Bank, 199 occurrences of the motif were found. This list has been saved to the file below.
When using TOMTOM to search the JASPAR Core database, one comparative motif was found. This molecule, ATHB-5, is found in Arabadosis thaliana. This is encouraging, because the Arabadopsis thaliana ortholog of the MT-CO1, was very similar to the human gene. It is possible the human ortholog was not within the database.
When using FIMO to search the Protein Data Bank, 199 occurrences of the motif were found. This list has been saved to the file below.
| fimo_results_motif_3.rtf |
YMF 3.0 Search

University of Washington - Computer Science & Engineering
The University of Washington - Computer Science & Engineering Department created the YMF motif finder. The actual algorithms used were unspecified, however, the default settings were used during the search.
The motifs were not extrapolated into functional data of the DNA or protein, and therefore need to be searched in other motif databases in order to determine their significance. Due to the large number of Motifs found (listed below), the searches were not completed.
The motifs were not extrapolated into functional data of the DNA or protein, and therefore need to be searched in other motif databases in order to determine their significance. Due to the large number of Motifs found (listed below), the searches were not completed.
Motif Count Zscore
AYSATA 12 13.46
TATSAW 12 12.07
AYCRTA 9 11.34
RTAGGW 14 10.75
TAGGAY 9 10.61
AYCATA 7 10.58
ATSATA 7 10.50
ATSRTA 9 10.37
TATCRW 8 10.27
TASGAY 10 10.24
TATGRW 11 10.22
ATCRTA 5 9.99
ATAGTR 7 9.92
ATAGGW 8 9.84
AYYATA 11 9.80
AYCAYA 13 9.73
ACYATA 7 9.70
RTAGTR 11 9.56
ATARTS 12 9.50
ATTSAT 7 9.39
ATCATA 4 9.35
TACWAY 10 9.32
ATGAWW 12 9.32
TATCAW 6 9.31
ATARWC 12 9.31
TACTAY 6 9.29
CTATSW 10 9.29
AYGATA 5 9.23
GTARTA 7 9.13
TAYCAW 9 9.08
ATAAWC 9 9.03
GTCCTA 6 9.01
TAGGWY 11 8.94
ATSRAT 9 8.94
TATGWW 9 8.93
TAYTAY 10 8.85
ATAGTS 7 8.82
ATAGTG 5 8.78
TCCTAY 9 8.74
ATSWAT 9 8.74
ACYAYA 12 8.74
TCSTAY 10 8.70
ACYRTA 9 8.68
TAGGWS 14 8.66
TATRGW 10 8.60
TAGGRY 13 8.60
TAYTAS 10 8.58
ACTATR 6 8.54
CCTAWY 12 8.53
ATSAWA 11 8.52
ACSATA 5 8.47
TATGAW 6 8.42
ATAGSW 10 8.41
ATARTC 7 8.40
TASTAY 9 8.39
WCCTAW 11 8.38
AYCSTA 8 8.38
CTATWC 7 8.37
TAYCWA 8 8.34
AWCRTA 7 8.33
CCTATY 8 8.31
AWSATA 9 8.31
ARTART 12 8.30
TAYGAW 7 8.28
ATAGYR 9 8.21
ATCAAT 4 8.19
CTAYWC 12 8.18
ATASGW 8 8.16
GTASTA 6 8.15
CTATSA 6 8.15
AYGATR 8 8.15
GTAGTA 4 8.14
ACSAYA 10 8.14
ATSAAW 11 8.13
CCTAYY 13 8.12
TAGGAS 10 8.11
ACYATW 10 8.10
TATRGY 10 8.07
ATYRAT 10 8.07
ATAATC 5 8.06
CSATAW 10 8.04
CTAYYA 10 8.00
TARGAY 10 7.95
TAGRAY 10 7.90
ASYATA 9 7.90
CTAYSA 9 7.87
CTAWYA 10 7.82
AYTATW 11 7.82
ACTATA 4 7.80
TATGRY 9 7.79
ATYAWT 12 7.79
ATAGGA 5 7.75
ATSAWT 10 7.74
AYGWTA 7 7.73
GTCSTA 6 7.70
ATCAWW 9 7.68
ATTRWT 12 7.63
TATSGW 7 7.62
CATAWW 11 7.61
AYGAWA 9 7.61
ATAGRW 9 7.61
ACYATR 9 7.60
WTGATR 9 7.59
ATSATW 10 7.59
ACTAYW 10 7.59
ATSWTA 8 7.58
GTWGTA 6 7.56
STAGGW 12 7.53
ATWATY 13 7.50
ATAWWC 11 7.50
ATRATY 10 7.48
TAGSAS 13 7.46
GWCCTA 7 7.46
TACYAY 8 7.43
TAGSAY 9 7.42
AYCATW 9 7.41
AYGATW 7 7.39
ACTATW 6 7.39
ACGWTR 7 7.39
ATSATR 10 7.37
ATGAWA 7 7.37
ACGWTA 4 7.36
TCYTAY 11 7.34
GTWRTA 9 7.34
ATGRWT 9 7.34
ATCRTR 7 7.34
RATAGR 10 7.33
ATAATY 7 7.32
ACSRTA 6 7.32
TATYAW 9 7.31
TACCTA 3 7.31
ATAGYG 6 7.31
CTAWSA 9 7.30
CTATWS 10 7.30
TAGGWR 12 7.29
AYCWTA 8 7.28
ASWATA 10 7.27
TYCTAY 11 7.26
ATRGTR 9 7.25
ATASSA 8 7.25
ATYATW 10 7.24
AWCATA 5 7.23
ATGYAT 6 7.23
ATSAYA 9 7.22
ATSART 9 7.21
TAGGWW 9 7.20
TACCWA 5 7.20
ATAGWR 9 7.19
TSCTAW 10 7.18
CCTAYS 10 7.18
ATRAWC 10 7.18
ATCAYA 6 7.16
CCTATS 6 7.15
AYGRTA 6 7.15
ATRYAT 10 7.14
ATAGWS 9 7.13
TAGGTR 6 7.12
GWATAR 10 7.12
ATWGTR 9 7.12
ATASTR 8 7.12
TWGGAY 10 7.09
GWATAS 9 7.09
AYCCTA 6 7.09
ATGAWR 11 7.09
CYATAY 9 7.08
CCTATC 4 7.08
ATRSAT 8 7.08
ATARTY 9 7.08
TATSWA 7 7.07
ACSWTA 7 7.07
ATCWTA 5 7.06
ATATSR 9 7.06
TARTAS 8 7.03
TSCTAY 10 7.02
TAGWSA 10 7.01
ACTAYR 8 7.01
TWTCAW 10 7.00
TATTAS 6 7.00
ATGRTW 8 7.00
WATCAW 8 6.98
ATAAWY 11 6.98
TAYGGW 7 6.96
RCTATR 8 6.95
TAWTAS 10 6.93
ATGSAT 6 6.93
TASGAS 10 6.91
ATAWTC 7 6.91
WTAGTR 8 6.90
TAGGRS 14 6.90
ATRRTC 9 6.90
WTATGR 10 6.89
CTTCGA 4 6.89
ATWAWC 10 6.89
ATRATC 6 6.89
TAYSAA 7 6.88
ATAGSA 6 6.88
CTAYGW 7 6.87
WATGAW 9 6.86
TAYCAA 5 6.86
TARTAR 9 6.86
ATGAAW 7 6.85
ATCSTA 4 6.82
GTASGW 8 6.80
GWCSTA 8 6.79
CTATGW 5 6.79
CTAYYC 11 6.78
ATCAYW 9 6.78
ATASGA 5 6.78
ATYSTA 7 6.77
WTCATR 9 6.76
TAYYAA 8 6.76
CTAYCA 6 6.76
ATWSGA 8 6.76
AATART 7 6.76
TTCGAA 4 6.75
ATCRYA 7 6.75
TRATAR 9 6.74
TATSRA 7 6.74
TATCWW 7 6.74
AYTATY 9 6.74
TCCTAW 7 6.73
TATSAA 5 6.73
ATGWAW 9 6.73
TAGRAS 12 6.72
CSTATY 8 6.72
SATAGS 8 6.71
ATARGW 9 6.70
ACCAYA 7 6.70
ATGATA 3 6.69
ATGWAT 5 6.67
AYCAWA 9 6.66
CCATAW 7 6.65
ATYAYT 10 6.65
TAGTSA 6 6.60
GWASTA 9 6.60
TAWCGW 5 6.59
ATCRTW 6 6.59
WCATAR 8 6.57
GTWSTA 8 6.57
ATTAYT 7 6.57
ATYRTA 7 6.56
ACGATA 2 6.56
TWGTAR 9 6.55
RTAGGR 11 6.54
GARTAR 11 6.54
CYTATY 10 6.54
GRATAR 10 6.52
CTATYC 6 6.52
ATRRTA 7 6.51
TATCAA 3 6.50
AYSATA 12 13.46
TATSAW 12 12.07
AYCRTA 9 11.34
RTAGGW 14 10.75
TAGGAY 9 10.61
AYCATA 7 10.58
ATSATA 7 10.50
ATSRTA 9 10.37
TATCRW 8 10.27
TASGAY 10 10.24
TATGRW 11 10.22
ATCRTA 5 9.99
ATAGTR 7 9.92
ATAGGW 8 9.84
AYYATA 11 9.80
AYCAYA 13 9.73
ACYATA 7 9.70
RTAGTR 11 9.56
ATARTS 12 9.50
ATTSAT 7 9.39
ATCATA 4 9.35
TACWAY 10 9.32
ATGAWW 12 9.32
TATCAW 6 9.31
ATARWC 12 9.31
TACTAY 6 9.29
CTATSW 10 9.29
AYGATA 5 9.23
GTARTA 7 9.13
TAYCAW 9 9.08
ATAAWC 9 9.03
GTCCTA 6 9.01
TAGGWY 11 8.94
ATSRAT 9 8.94
TATGWW 9 8.93
TAYTAY 10 8.85
ATAGTS 7 8.82
ATAGTG 5 8.78
TCCTAY 9 8.74
ATSWAT 9 8.74
ACYAYA 12 8.74
TCSTAY 10 8.70
ACYRTA 9 8.68
TAGGWS 14 8.66
TATRGW 10 8.60
TAGGRY 13 8.60
TAYTAS 10 8.58
ACTATR 6 8.54
CCTAWY 12 8.53
ATSAWA 11 8.52
ACSATA 5 8.47
TATGAW 6 8.42
ATAGSW 10 8.41
ATARTC 7 8.40
TASTAY 9 8.39
WCCTAW 11 8.38
AYCSTA 8 8.38
CTATWC 7 8.37
TAYCWA 8 8.34
AWCRTA 7 8.33
CCTATY 8 8.31
AWSATA 9 8.31
ARTART 12 8.30
TAYGAW 7 8.28
ATAGYR 9 8.21
ATCAAT 4 8.19
CTAYWC 12 8.18
ATASGW 8 8.16
GTASTA 6 8.15
CTATSA 6 8.15
AYGATR 8 8.15
GTAGTA 4 8.14
ACSAYA 10 8.14
ATSAAW 11 8.13
CCTAYY 13 8.12
TAGGAS 10 8.11
ACYATW 10 8.10
TATRGY 10 8.07
ATYRAT 10 8.07
ATAATC 5 8.06
CSATAW 10 8.04
CTAYYA 10 8.00
TARGAY 10 7.95
TAGRAY 10 7.90
ASYATA 9 7.90
CTAYSA 9 7.87
CTAWYA 10 7.82
AYTATW 11 7.82
ACTATA 4 7.80
TATGRY 9 7.79
ATYAWT 12 7.79
ATAGGA 5 7.75
ATSAWT 10 7.74
AYGWTA 7 7.73
GTCSTA 6 7.70
ATCAWW 9 7.68
ATTRWT 12 7.63
TATSGW 7 7.62
CATAWW 11 7.61
AYGAWA 9 7.61
ATAGRW 9 7.61
ACYATR 9 7.60
WTGATR 9 7.59
ATSATW 10 7.59
ACTAYW 10 7.59
ATSWTA 8 7.58
GTWGTA 6 7.56
STAGGW 12 7.53
ATWATY 13 7.50
ATAWWC 11 7.50
ATRATY 10 7.48
TAGSAS 13 7.46
GWCCTA 7 7.46
TACYAY 8 7.43
TAGSAY 9 7.42
AYCATW 9 7.41
AYGATW 7 7.39
ACTATW 6 7.39
ACGWTR 7 7.39
ATSATR 10 7.37
ATGAWA 7 7.37
ACGWTA 4 7.36
TCYTAY 11 7.34
GTWRTA 9 7.34
ATGRWT 9 7.34
ATCRTR 7 7.34
RATAGR 10 7.33
ATAATY 7 7.32
ACSRTA 6 7.32
TATYAW 9 7.31
TACCTA 3 7.31
ATAGYG 6 7.31
CTAWSA 9 7.30
CTATWS 10 7.30
TAGGWR 12 7.29
AYCWTA 8 7.28
ASWATA 10 7.27
TYCTAY 11 7.26
ATRGTR 9 7.25
ATASSA 8 7.25
ATYATW 10 7.24
AWCATA 5 7.23
ATGYAT 6 7.23
ATSAYA 9 7.22
ATSART 9 7.21
TAGGWW 9 7.20
TACCWA 5 7.20
ATAGWR 9 7.19
TSCTAW 10 7.18
CCTAYS 10 7.18
ATRAWC 10 7.18
ATCAYA 6 7.16
CCTATS 6 7.15
AYGRTA 6 7.15
ATRYAT 10 7.14
ATAGWS 9 7.13
TAGGTR 6 7.12
GWATAR 10 7.12
ATWGTR 9 7.12
ATASTR 8 7.12
TWGGAY 10 7.09
GWATAS 9 7.09
AYCCTA 6 7.09
ATGAWR 11 7.09
CYATAY 9 7.08
CCTATC 4 7.08
ATRSAT 8 7.08
ATARTY 9 7.08
TATSWA 7 7.07
ACSWTA 7 7.07
ATCWTA 5 7.06
ATATSR 9 7.06
TARTAS 8 7.03
TSCTAY 10 7.02
TAGWSA 10 7.01
ACTAYR 8 7.01
TWTCAW 10 7.00
TATTAS 6 7.00
ATGRTW 8 7.00
WATCAW 8 6.98
ATAAWY 11 6.98
TAYGGW 7 6.96
RCTATR 8 6.95
TAWTAS 10 6.93
ATGSAT 6 6.93
TASGAS 10 6.91
ATAWTC 7 6.91
WTAGTR 8 6.90
TAGGRS 14 6.90
ATRRTC 9 6.90
WTATGR 10 6.89
CTTCGA 4 6.89
ATWAWC 10 6.89
ATRATC 6 6.89
TAYSAA 7 6.88
ATAGSA 6 6.88
CTAYGW 7 6.87
WATGAW 9 6.86
TAYCAA 5 6.86
TARTAR 9 6.86
ATGAAW 7 6.85
ATCSTA 4 6.82
GTASGW 8 6.80
GWCSTA 8 6.79
CTATGW 5 6.79
CTAYYC 11 6.78
ATCAYW 9 6.78
ATASGA 5 6.78
ATYSTA 7 6.77
WTCATR 9 6.76
TAYYAA 8 6.76
CTAYCA 6 6.76
ATWSGA 8 6.76
AATART 7 6.76
TTCGAA 4 6.75
ATCRYA 7 6.75
TRATAR 9 6.74
TATSRA 7 6.74
TATCWW 7 6.74
AYTATY 9 6.74
TCCTAW 7 6.73
TATSAA 5 6.73
ATGWAW 9 6.73
TAGRAS 12 6.72
CSTATY 8 6.72
SATAGS 8 6.71
ATARGW 9 6.70
ACCAYA 7 6.70
ATGATA 3 6.69
ATGWAT 5 6.67
AYCAWA 9 6.66
CCATAW 7 6.65
ATYAYT 10 6.65
TAGTSA 6 6.60
GWASTA 9 6.60
TAWCGW 5 6.59
ATCRTW 6 6.59
WCATAR 8 6.57
GTWSTA 8 6.57
ATTAYT 7 6.57
ATYRTA 7 6.56
ACGATA 2 6.56
TWGTAR 9 6.55
RTAGGR 11 6.54
GARTAR 11 6.54
CYTATY 10 6.54
GRATAR 10 6.52
CTATYC 6 6.52
ATRRTA 7 6.51
TATCAA 3 6.50
Motif Count Zscore
AWCSTA 6 6.48
ATCRAT 4 6.48
TCCTAS 8 6.47
GTRRTA 8 6.47
CTAWCW 8 6.47
ATGATW 5 6.46
ATCATW 5 6.46
GWTAGR 8 6.45
CSTAGR 12 6.45
AYAAWC 10 6.45
AWTAGG 7 6.45
ATCATR 5 6.45
TSATAR 8 6.44
AYTATR 8 6.44
ACCRTA 4 6.44
AATATR 7 6.44
TCSTAS 9 6.42
TAGTRR 9 6.41
RATAGS 9 6.41
ATWGGW 9 6.40
ATTSWT 9 6.40
ATAGRA 6 6.40
TATSAY 7 6.39
GTYSTA 8 6.38
ATAWTS 10 6.38
CTATCW 5 6.37
AYCATR 8 6.37
ARTASG 9 6.37
TAWGAW 8 6.36
TATSGY 7 6.36
TASGTR 7 6.35
CTAYTC 7 6.35
ATAAYC 6 6.35
CATATW 6 6.34
ATAGWG 6 6.34
ASSATA 7 6.34
GTARTW 8 6.33
GATAGR 5 6.33
GAATAR 7 6.33
ATAWTY 11 6.33
RTAGTW 7 6.32
ATGRAT 5 6.32
TWCTAY 8 6.31
GTSCTA 6 6.30
CGAWWA 8 6.28
RTGAAW 11 6.27
GATAGS 5 6.27
ACCTAY 5 6.27
ATATGR 6 6.26
AATAAT 5 6.26
AWTATG 6 6.25
ATYATY 9 6.25
CTATYA 5 6.23
TRATAS 7 6.22
TRTTAS 8 6.21
ATWATC 6 6.21
ATTSAY 8 6.21
GTYCTA 6 6.20
CYATWC 9 6.20
TATGGW 5 6.19
TARTAY 7 6.19
CATAWY 8 6.19
ATRGTS 9 6.19
ACWATR 8 6.19
ATARTR 8 6.18
AATAYR 9 6.18
GTCYTA 6 6.17
AWAATC 8 6.17
ATAGYS 8 6.17
ACYAWA 9 6.17
ACCRYA 9 6.17
GAYTAY 8 6.16
ATWGRT 7 6.16
TARTAW 8 6.15
ATSCRT 7 6.15
SCATAW 9 6.14
GAWTAW 10 6.14
TATASY 8 6.13
GYARTA 9 6.13
GWTTAY 9 6.13
ATGATR 5 6.13
ASTATR 7 6.13
ACWAYA 9 6.13
TARGAS 11 6.12
GTATAS 5 6.12
ATTSRT 7 6.12
ATRWTC 9 6.12
ATAYGR 7 6.12
TATWGW 8 6.11
CTWCGW 7 6.11
AWCWTA 7 6.11
GTATRS 7 6.10
CWSCTA 12 6.10
CTAWCA 5 6.10
ATRGGW 10 6.10
ASTATW 8 6.09
ASTATA 5 6.09
ATCAWT 5 6.08
ATCART 5 6.08
ACCTAT 3 6.08
TWTCAY 10 6.07
CYTATS 8 6.07
WCTATR 7 6.06
TRCTAY 7 6.05
ATGSWT 8 6.05
GTAGTW 5 6.04
CTATAC 3 6.04
TACTRY 8 6.03
AYRATA 7 6.03
WATAGW 8 6.02
CGATWA 4 6.02
ATRATW 8 6.02
TWCGAW 5 6.01
ACTRTA 5 6.01
TCYTAS 10 6.00
TATYAR 8 6.00
TATAGW 5 6.00
CWAYAC 9 6.00
CCRTAW 8 6.00
AYACSA 8 6.00
TATYRA 7 5.98
CTWYGA 8 5.98
ATGCAT 4 5.98
ACSTAY 6 5.98
ATGRTA 4 5.97
WTAGGR 10 5.96
TWATCR 7 5.96
CGWWGA 8 5.96
AATWRT 10 5.96
TAWTCW 9 5.95
CGAWRA 9 5.95
ATAYGG 5 5.95
ATATSG 5 5.95
TASTAS 8 5.94
CTATTC 4 5.93
ATGWWT 8 5.93
ACSTAS 7 5.93
TATGSW 7 5.92
GTRGTA 5 5.92
CTATCA 3 5.92
ATARGA 6 5.92
TATAGY 5 5.91
TAGTRW 7 5.90
AWARTC 11 5.90
ACCATA 3 5.90
AATRRT 9 5.90
AYACCW 10 5.89
ATCRRT 6 5.89
TCCWAY 10 5.88
RTAGTS 8 5.88
CWATAY 8 5.88
TAGYRA 7 5.87
STAGGR 12 5.87
ASRATA 8 5.87
TARTCR 7 5.86
CWATWC 8 5.86
CTAWYC 8 5.86
CGAWTA 4 5.86
AWYCTA 7 5.86
TAWTCR 7 5.85
TAGRTR 7 5.84
CCTASS 12 5.84
ATGRAW 9 5.84
TAGTRA 5 5.83
TACYAW 6 5.83
TACWAW 7 5.83
TGATRR 9 5.82
SATAGR 7 5.81
CRTATW 7 5.81
ATAGGR 6 5.81
ARTAGG 7 5.81
CSTATC 4 5.80
ATGAWT 5 5.80
ATASRA 7 5.80
ATAGSR 8 5.80
TTSATR 8 5.79
ATTATY 6 5.79
ATAYYA 7 5.79
ACTRTW 8 5.79
RTGATR 8 5.78
GTWGTW 8 5.78
TTCGAW 4 5.77
CTAWWC 8 5.77
CSTATS 6 5.77
AYAGTR 8 5.77
ATASGR 7 5.77
GTAGGW 6 5.76
AWYATA 8 5.76
ATATGG 4 5.76
TCSTAW 7 5.75
ATSAAY 7 5.75
ATCWAT 4 5.75
TTCGWA 4 5.74
CGATRW 6 5.74
ACWATW 8 5.74
TACYWA 6 5.73
AWTATR 9 5.73
ATYCRT 6 5.73
ATATRR 9 5.73
CYATCA 6 5.72
ATARYC 8 5.72
ACWATA 5 5.72
GATARS 7 5.71
CTTCGW 5 5.71
ATCCTA 3 5.71
TAYGAY 5 5.70
TASRTA 5 5.70
CYTATC 5 5.70
ATATYR 8 5.70
TARTSA 8 5.69
ATWTSA 9 5.69
ATRGTG 6 5.69
AATATG 4 5.69
ATWRTC 8 5.68
TATYAA 5 5.67
CTAYCW 8 5.67
CCTAWS 8 5.67
ATRAWA 10 5.67
CTRYTA 9 5.66
CCTAYC 6 5.66
ATTGWT 5 5.66
TAWTAR 10 5.65
TAGGTW 4 5.65
CTASGW 8 5.65
ATRATS 8 5.65
GATASR 6 5.64
TAGWCA 5 5.63
TAGRTW 6 5.63
GTASGA 5 5.63
CTATGA 3 5.63
ATWGGA 6 5.63
ATASYA 6 5.63
ACACSW 10 5.63
AATRAT 6 5.63
TAGYAR 8 5.62
TACTWY 8 5.62
GAATRR 10 5.62
TATYAY 7 5.61
GAWGAW 12 5.61
ATWGTA 4 5.61
ATRCAW 7 5.61
ATGYRT 7 5.61
ATCAWY 7 5.61
AAYART 9 5.61
YGATAR 6 5.60
TSATAS 6 5.60
RTATTR 8 5.60
ACTAYA 5 5.60
AATWAT 7 5.60
TTCGAR 5 5.59
GTWGWA 8 5.59
AWCSTA 6 6.48
ATCRAT 4 6.48
TCCTAS 8 6.47
GTRRTA 8 6.47
CTAWCW 8 6.47
ATGATW 5 6.46
ATCATW 5 6.46
GWTAGR 8 6.45
CSTAGR 12 6.45
AYAAWC 10 6.45
AWTAGG 7 6.45
ATCATR 5 6.45
TSATAR 8 6.44
AYTATR 8 6.44
ACCRTA 4 6.44
AATATR 7 6.44
TCSTAS 9 6.42
TAGTRR 9 6.41
RATAGS 9 6.41
ATWGGW 9 6.40
ATTSWT 9 6.40
ATAGRA 6 6.40
TATSAY 7 6.39
GTYSTA 8 6.38
ATAWTS 10 6.38
CTATCW 5 6.37
AYCATR 8 6.37
ARTASG 9 6.37
TAWGAW 8 6.36
TATSGY 7 6.36
TASGTR 7 6.35
CTAYTC 7 6.35
ATAAYC 6 6.35
CATATW 6 6.34
ATAGWG 6 6.34
ASSATA 7 6.34
GTARTW 8 6.33
GATAGR 5 6.33
GAATAR 7 6.33
ATAWTY 11 6.33
RTAGTW 7 6.32
ATGRAT 5 6.32
TWCTAY 8 6.31
GTSCTA 6 6.30
CGAWWA 8 6.28
RTGAAW 11 6.27
GATAGS 5 6.27
ACCTAY 5 6.27
ATATGR 6 6.26
AATAAT 5 6.26
AWTATG 6 6.25
ATYATY 9 6.25
CTATYA 5 6.23
TRATAS 7 6.22
TRTTAS 8 6.21
ATWATC 6 6.21
ATTSAY 8 6.21
GTYCTA 6 6.20
CYATWC 9 6.20
TATGGW 5 6.19
TARTAY 7 6.19
CATAWY 8 6.19
ATRGTS 9 6.19
ACWATR 8 6.19
ATARTR 8 6.18
AATAYR 9 6.18
GTCYTA 6 6.17
AWAATC 8 6.17
ATAGYS 8 6.17
ACYAWA 9 6.17
ACCRYA 9 6.17
GAYTAY 8 6.16
ATWGRT 7 6.16
TARTAW 8 6.15
ATSCRT 7 6.15
SCATAW 9 6.14
GAWTAW 10 6.14
TATASY 8 6.13
GYARTA 9 6.13
GWTTAY 9 6.13
ATGATR 5 6.13
ASTATR 7 6.13
ACWAYA 9 6.13
TARGAS 11 6.12
GTATAS 5 6.12
ATTSRT 7 6.12
ATRWTC 9 6.12
ATAYGR 7 6.12
TATWGW 8 6.11
CTWCGW 7 6.11
AWCWTA 7 6.11
GTATRS 7 6.10
CWSCTA 12 6.10
CTAWCA 5 6.10
ATRGGW 10 6.10
ASTATW 8 6.09
ASTATA 5 6.09
ATCAWT 5 6.08
ATCART 5 6.08
ACCTAT 3 6.08
TWTCAY 10 6.07
CYTATS 8 6.07
WCTATR 7 6.06
TRCTAY 7 6.05
ATGSWT 8 6.05
GTAGTW 5 6.04
CTATAC 3 6.04
TACTRY 8 6.03
AYRATA 7 6.03
WATAGW 8 6.02
CGATWA 4 6.02
ATRATW 8 6.02
TWCGAW 5 6.01
ACTRTA 5 6.01
TCYTAS 10 6.00
TATYAR 8 6.00
TATAGW 5 6.00
CWAYAC 9 6.00
CCRTAW 8 6.00
AYACSA 8 6.00
TATYRA 7 5.98
CTWYGA 8 5.98
ATGCAT 4 5.98
ACSTAY 6 5.98
ATGRTA 4 5.97
WTAGGR 10 5.96
TWATCR 7 5.96
CGWWGA 8 5.96
AATWRT 10 5.96
TAWTCW 9 5.95
CGAWRA 9 5.95
ATAYGG 5 5.95
ATATSG 5 5.95
TASTAS 8 5.94
CTATTC 4 5.93
ATGWWT 8 5.93
ACSTAS 7 5.93
TATGSW 7 5.92
GTRGTA 5 5.92
CTATCA 3 5.92
ATARGA 6 5.92
TATAGY 5 5.91
TAGTRW 7 5.90
AWARTC 11 5.90
ACCATA 3 5.90
AATRRT 9 5.90
AYACCW 10 5.89
ATCRRT 6 5.89
TCCWAY 10 5.88
RTAGTS 8 5.88
CWATAY 8 5.88
TAGYRA 7 5.87
STAGGR 12 5.87
ASRATA 8 5.87
TARTCR 7 5.86
CWATWC 8 5.86
CTAWYC 8 5.86
CGAWTA 4 5.86
AWYCTA 7 5.86
TAWTCR 7 5.85
TAGRTR 7 5.84
CCTASS 12 5.84
ATGRAW 9 5.84
TAGTRA 5 5.83
TACYAW 6 5.83
TACWAW 7 5.83
TGATRR 9 5.82
SATAGR 7 5.81
CRTATW 7 5.81
ATAGGR 6 5.81
ARTAGG 7 5.81
CSTATC 4 5.80
ATGAWT 5 5.80
ATASRA 7 5.80
ATAGSR 8 5.80
TTSATR 8 5.79
ATTATY 6 5.79
ATAYYA 7 5.79
ACTRTW 8 5.79
RTGATR 8 5.78
GTWGTW 8 5.78
TTCGAW 4 5.77
CTAWWC 8 5.77
CSTATS 6 5.77
AYAGTR 8 5.77
ATASGR 7 5.77
GTAGGW 6 5.76
AWYATA 8 5.76
ATATGG 4 5.76
TCSTAW 7 5.75
ATSAAY 7 5.75
ATCWAT 4 5.75
TTCGWA 4 5.74
CGATRW 6 5.74
ACWATW 8 5.74
TACYWA 6 5.73
AWTATR 9 5.73
ATYCRT 6 5.73
ATATRR 9 5.73
CYATCA 6 5.72
ATARYC 8 5.72
ACWATA 5 5.72
GATARS 7 5.71
CTTCGW 5 5.71
ATCCTA 3 5.71
TAYGAY 5 5.70
TASRTA 5 5.70
CYTATC 5 5.70
ATATYR 8 5.70
TARTSA 8 5.69
ATWTSA 9 5.69
ATRGTG 6 5.69
AATATG 4 5.69
ATWRTC 8 5.68
TATYAA 5 5.67
CTAYCW 8 5.67
CCTAWS 8 5.67
ATRAWA 10 5.67
CTRYTA 9 5.66
CCTAYC 6 5.66
ATTGWT 5 5.66
TAWTAR 10 5.65
TAGGTW 4 5.65
CTASGW 8 5.65
ATRATS 8 5.65
GATASR 6 5.64
TAGWCA 5 5.63
TAGRTW 6 5.63
GTASGA 5 5.63
CTATGA 3 5.63
ATWGGA 6 5.63
ATASYA 6 5.63
ACACSW 10 5.63
AATRAT 6 5.63
TAGYAR 8 5.62
TACTWY 8 5.62
GAATRR 10 5.62
TATYAY 7 5.61
GAWGAW 12 5.61
ATWGTA 4 5.61
ATRCAW 7 5.61
ATGYRT 7 5.61
ATCAWY 7 5.61
AAYART 9 5.61
YGATAR 6 5.60
TSATAS 6 5.60
RTATTR 8 5.60
ACTAYA 5 5.60
AATWAT 7 5.60
TTCGAR 5 5.59
GTWGWA 8 5.59
Motif Count Zscore
GTAWTA 5 5.59
GATTAY 5 5.59
CYATSA 9 5.59
ACRTWG 7 5.59
ATTRAT 6 5.58
TARTCA 5 5.57
TRTAGW 8 5.56
TATGAY 4 5.56
GTRSTA 7 5.56
ATTRYT 9 5.56
ACRTAG 4 5.56
GTAYTA 4 5.55
ATRCRT 6 5.55
TATWCY 9 5.54
TATGGY 5 5.54
CCTAWC 5 5.54
ATASTG 5 5.53
TARTRA 8 5.52
GYCCTA 7 5.52
GTASTW 7 5.52
CTAAYA 5 5.52
CATATY 5 5.52
GGWRTA 10 5.51
AYCYTA 7 5.51
AWTART 9 5.51
ACGATR 3 5.51
RATATR 8 5.50
GTARGW 8 5.50
TRGTAS 7 5.49
TACSWA 5 5.49
TAGTSR 8 5.48
ATWTCA 6 5.48
ATAAYY 8 5.48
ACSATW 6 5.48
ACCRWA 7 5.48
TRTAGY 8 5.47
TATGWA 4 5.47
TRATAW 8 5.46
RTATGS 7 5.46
GWARTA 9 5.46
CCAYAW 11 5.46
AYARTC 8 5.46
AWGATR 8 5.46
ATWSTA 6 5.46
ARTATR 8 5.46
CTATYS 7 5.45
ATWCAY 7 5.45
WCCTAR 9 5.44
TARWCA 8 5.44
ATTAWT 7 5.44
GTARYA 7 5.43
AYGAYA 6 5.43
CSATAA 5 5.42
ATGRWA 8 5.42
ATARYG 7 5.41
ACSAWA 7 5.41
TAGSTR 8 5.40
RTATGR 7 5.40
ATTCGA 2 5.40
ATASST 6 5.40
ACSATR 6 5.40
TACCWW 6 5.39
GTSSTA 7 5.39
TATGWY 6 5.38
GTYTAY 8 5.38
CTATWY 8 5.38
CCTASG 8 5.38
CCATAY 5 5.38
CATAYW 7 5.38
AWAGTR 9 5.38
ATWAYT 9 5.38
ATRCWT 7 5.38
YCCTAR 11 5.37
TAGTAS 4 5.37
GWAGWA 11 5.37
CTATSR 6 5.37
AWGAWA 10 5.37
ATTSAW 7 5.37
AGAWTA 6 5.37
CGATRA 4 5.36
AYYCTA 7 5.36
ATRCAY 7 5.36
TRTGAW 8 5.35
TASGTW 5 5.35
GTCCWA 6 5.35
CWYCTA 10 5.35
ATRGSA 9 5.35
ACRTAR 6 5.35
TTCGRA 5 5.34
ASCAYA 9 5.34
ACGTWR 6 5.34
CAWCAY 9 5.33
AAYAAT 6 5.33
TATCRA 3 5.32
RTCCTW 9 5.32
CTWCGA 4 5.32
CTATAS 5 5.32
ATYATR 7 5.32
ATGAAT 3 5.32
ATAYSA 6 5.32
AWAAWC 12 5.31
ATSCAW 7 5.31
ATASTS 7 5.31
GWCTAY 7 5.30
CSATAY 6 5.30
ATTAYY 8 5.30
TWCGRA 6 5.29
TAYCRA 5 5.29
TAWCAW 6 5.29
GWSCTA 8 5.29
CWTCGW 6 5.29
ATGWRT 7 5.29
ATSSTA 5 5.28
ACCSTA 4 5.28
WTATCR 6 5.27
TYCTAS 10 5.27
TCCYAY 12 5.27
TAYACS 7 5.27
GRTAGR 8 5.27
ATTGWW 7 5.27
ATAYSG 6 5.27
TGATWW 8 5.26
TASGAW 6 5.26
GWTTAW 9 5.26
CTAYTS 9 5.26
CACTAW 5 5.26
AYTRTA 7 5.26
AYCCWA 8 5.26
ATCAYR 7 5.26
ACRAYA 7 5.26
TATTSW 8 5.25
TAGSAR 10 5.25
CYAWCA 9 5.25
CWTCGA 4 5.25
CTATYY 8 5.25
CCTASR 11 5.25
ACCRTW 6 5.25
ACACSA 6 5.25
ACACGA 3 5.25
TARTAA 5 5.24
GTWGTR 8 5.24
GTAGRW 8 5.24
CYATCW 9 5.24
AYAATY 8 5.24
ATCRWA 5 5.24
TWGTAS 7 5.23
TCGAAS 4 5.23
TATASW 7 5.23
GTCSWA 7 5.23
GRATAS 7 5.23
CYTAYC 8 5.23
CWCCTA 7 5.23
CTAWTC 5 5.23
AYGAAW 7 5.23
TRCTAW 6 5.22
TAGGWA 5 5.22
GYATAS 6 5.22
CTACYA 5 5.22
TYATCR 7 5.21
TWGGTR 8 5.21
GRTAGS 8 5.21
GGTRTW 9 5.21
ATARTG 5 5.21
YCTAGR 12 5.20
TATTRS 8 5.20
GTAGWA 5 5.20
AYSCTA 6 5.20
AYCTAY 6 5.20
ATCAAY 4 5.20
ACTAYY 7 5.20
GTASWA 7 5.19
GATWAY 7 5.19
AYAATC 5 5.19
ATAGRR 8 5.19
TATRAW 8 5.18
ACTAYT 5 5.18
AYWATC 7 5.17
ATAAWS 9 5.17
TSCTAR 9 5.16
TASCWA 6 5.16
CTRTWC 9 5.16
CTASGR 9 5.16
CCWATY 9 5.16
ATAWYC 8 5.16
TAWYAA 8 5.15
TATYWA 7 5.15
CTAYAC 5 5.15
CACTAY 5 5.15
AYAWTC 8 5.15
ATTRTW 8 5.15
ATGSRT 7 5.15
ATATGS 5 5.15
ATACYW 6 5.15
TWGTRA 8 5.14
TACTAS 4 5.14
CYATAW 8 5.14
ACGTTR 4 5.14
TATTRR 8 5.13
TACSTA 3 5.13
SACTAW 7 5.13
CTATRY 6 5.13
AYTATS 6 5.13
ATGWWA 7 5.13
ARTAGR 9 5.13
TASTAW 6 5.12
GTARTR 7 5.12
CSTAGS 10 5.12
CYATAC 4 5.11
ATAWSG 7 5.11
ATASRT 5 5.11
GAYGAW 7 5.10
ACYWTA 7 5.10
GTAATA 3 5.09
AYRCCA 9 5.09
AWWTCA 9 5.09
ATCSWA 5 5.09
ACGWWG 6 5.09
TYCGAW 6 5.08
TRGTAR 7 5.08
TCGWAS 5 5.08
TAGYSA 7 5.08
GTAGWW 7 5.08
ATGRTR 7 5.08
ASCATA 4 5.08
ACWCSA 9 5.08
TATWCS 7 5.07
TAGRYA 6 5.07
CTAWTA 5 5.07
AWATSA 9 5.07
ATWCRT 5 5.07
ATRRTG 8 5.07
TTCGRW 6 5.06
CATARW 7 5.06
AWWATC 9 5.06
AWTASG 7 5.06
ATAAAC 4 5.06
AATAGG 4 5.06
WATATR 9 5.05
TYCTAW 8 5.05
TSCTAS 9 5.05
RCTATW 7 5.05
ARTAAY 8 5.05
AAYATR 8 5.05
GWYCTA 7 5.03
CYATYA 8 5.03
AWGATA 4 5.03
AWCCTA 4 5.03
ATCYTA 4 5.03
ASATAG 4 5.03
ACWRTA 7 5.03
TYCGRA 10 5.02
GTAWTA 5 5.59
GATTAY 5 5.59
CYATSA 9 5.59
ACRTWG 7 5.59
ATTRAT 6 5.58
TARTCA 5 5.57
TRTAGW 8 5.56
TATGAY 4 5.56
GTRSTA 7 5.56
ATTRYT 9 5.56
ACRTAG 4 5.56
GTAYTA 4 5.55
ATRCRT 6 5.55
TATWCY 9 5.54
TATGGY 5 5.54
CCTAWC 5 5.54
ATASTG 5 5.53
TARTRA 8 5.52
GYCCTA 7 5.52
GTASTW 7 5.52
CTAAYA 5 5.52
CATATY 5 5.52
GGWRTA 10 5.51
AYCYTA 7 5.51
AWTART 9 5.51
ACGATR 3 5.51
RATATR 8 5.50
GTARGW 8 5.50
TRGTAS 7 5.49
TACSWA 5 5.49
TAGTSR 8 5.48
ATWTCA 6 5.48
ATAAYY 8 5.48
ACSATW 6 5.48
ACCRWA 7 5.48
TRTAGY 8 5.47
TATGWA 4 5.47
TRATAW 8 5.46
RTATGS 7 5.46
GWARTA 9 5.46
CCAYAW 11 5.46
AYARTC 8 5.46
AWGATR 8 5.46
ATWSTA 6 5.46
ARTATR 8 5.46
CTATYS 7 5.45
ATWCAY 7 5.45
WCCTAR 9 5.44
TARWCA 8 5.44
ATTAWT 7 5.44
GTARYA 7 5.43
AYGAYA 6 5.43
CSATAA 5 5.42
ATGRWA 8 5.42
ATARYG 7 5.41
ACSAWA 7 5.41
TAGSTR 8 5.40
RTATGR 7 5.40
ATTCGA 2 5.40
ATASST 6 5.40
ACSATR 6 5.40
TACCWW 6 5.39
GTSSTA 7 5.39
TATGWY 6 5.38
GTYTAY 8 5.38
CTATWY 8 5.38
CCTASG 8 5.38
CCATAY 5 5.38
CATAYW 7 5.38
AWAGTR 9 5.38
ATWAYT 9 5.38
ATRCWT 7 5.38
YCCTAR 11 5.37
TAGTAS 4 5.37
GWAGWA 11 5.37
CTATSR 6 5.37
AWGAWA 10 5.37
ATTSAW 7 5.37
AGAWTA 6 5.37
CGATRA 4 5.36
AYYCTA 7 5.36
ATRCAY 7 5.36
TRTGAW 8 5.35
TASGTW 5 5.35
GTCCWA 6 5.35
CWYCTA 10 5.35
ATRGSA 9 5.35
ACRTAR 6 5.35
TTCGRA 5 5.34
ASCAYA 9 5.34
ACGTWR 6 5.34
CAWCAY 9 5.33
AAYAAT 6 5.33
TATCRA 3 5.32
RTCCTW 9 5.32
CTWCGA 4 5.32
CTATAS 5 5.32
ATYATR 7 5.32
ATGAAT 3 5.32
ATAYSA 6 5.32
AWAAWC 12 5.31
ATSCAW 7 5.31
ATASTS 7 5.31
GWCTAY 7 5.30
CSATAY 6 5.30
ATTAYY 8 5.30
TWCGRA 6 5.29
TAYCRA 5 5.29
TAWCAW 6 5.29
GWSCTA 8 5.29
CWTCGW 6 5.29
ATGWRT 7 5.29
ATSSTA 5 5.28
ACCSTA 4 5.28
WTATCR 6 5.27
TYCTAS 10 5.27
TCCYAY 12 5.27
TAYACS 7 5.27
GRTAGR 8 5.27
ATTGWW 7 5.27
ATAYSG 6 5.27
TGATWW 8 5.26
TASGAW 6 5.26
GWTTAW 9 5.26
CTAYTS 9 5.26
CACTAW 5 5.26
AYTRTA 7 5.26
AYCCWA 8 5.26
ATCAYR 7 5.26
ACRAYA 7 5.26
TATTSW 8 5.25
TAGSAR 10 5.25
CYAWCA 9 5.25
CWTCGA 4 5.25
CTATYY 8 5.25
CCTASR 11 5.25
ACCRTW 6 5.25
ACACSA 6 5.25
ACACGA 3 5.25
TARTAA 5 5.24
GTWGTR 8 5.24
GTAGRW 8 5.24
CYATCW 9 5.24
AYAATY 8 5.24
ATCRWA 5 5.24
TWGTAS 7 5.23
TCGAAS 4 5.23
TATASW 7 5.23
GTCSWA 7 5.23
GRATAS 7 5.23
CYTAYC 8 5.23
CWCCTA 7 5.23
CTAWTC 5 5.23
AYGAAW 7 5.23
TRCTAW 6 5.22
TAGGWA 5 5.22
GYATAS 6 5.22
CTACYA 5 5.22
TYATCR 7 5.21
TWGGTR 8 5.21
GRTAGS 8 5.21
GGTRTW 9 5.21
ATARTG 5 5.21
YCTAGR 12 5.20
TATTRS 8 5.20
GTAGWA 5 5.20
AYSCTA 6 5.20
AYCTAY 6 5.20
ATCAAY 4 5.20
ACTAYY 7 5.20
GTASWA 7 5.19
GATWAY 7 5.19
AYAATC 5 5.19
ATAGRR 8 5.19
TATRAW 8 5.18
ACTAYT 5 5.18
AYWATC 7 5.17
ATAAWS 9 5.17
TSCTAR 9 5.16
TASCWA 6 5.16
CTRTWC 9 5.16
CTASGR 9 5.16
CCWATY 9 5.16
ATAWYC 8 5.16
TAWYAA 8 5.15
TATYWA 7 5.15
CTAYAC 5 5.15
CACTAY 5 5.15
AYAWTC 8 5.15
ATTRTW 8 5.15
ATGSRT 7 5.15
ATATGS 5 5.15
ATACYW 6 5.15
TWGTRA 8 5.14
TACTAS 4 5.14
CYATAW 8 5.14
ACGTTR 4 5.14
TATTRR 8 5.13
TACSTA 3 5.13
SACTAW 7 5.13
CTATRY 6 5.13
AYTATS 6 5.13
ATGWWA 7 5.13
ARTAGR 9 5.13
TASTAW 6 5.12
GTARTR 7 5.12
CSTAGS 10 5.12
CYATAC 4 5.11
ATAWSG 7 5.11
ATASRT 5 5.11
GAYGAW 7 5.10
ACYWTA 7 5.10
GTAATA 3 5.09
AYRCCA 9 5.09
AWWTCA 9 5.09
ATCSWA 5 5.09
ACGWWG 6 5.09
TYCGAW 6 5.08
TRGTAR 7 5.08
TCGWAS 5 5.08
TAGYSA 7 5.08
GTAGWW 7 5.08
ATGRTR 7 5.08
ASCATA 4 5.08
ACWCSA 9 5.08
TATWCS 7 5.07
TAGRYA 6 5.07
CTAWTA 5 5.07
AWATSA 9 5.07
ATWCRT 5 5.07
ATRRTG 8 5.07
TTCGRW 6 5.06
CATARW 7 5.06
AWWATC 9 5.06
AWTASG 7 5.06
ATAAAC 4 5.06
AATAGG 4 5.06
WATATR 9 5.05
TYCTAW 8 5.05
TSCTAS 9 5.05
RCTATW 7 5.05
ARTAAY 8 5.05
AAYATR 8 5.05
GWYCTA 7 5.03
CYATYA 8 5.03
AWGATA 4 5.03
AWCCTA 4 5.03
ATCYTA 4 5.03
ASATAG 4 5.03
ACWRTA 7 5.03
TYCGRA 10 5.02
Motif Count Zscore
TATTAR 5 5.02
CRTAWA 7 5.02
AWAATY 11 5.02
ATWTYA 9 5.02
AGTART 5 5.02
AWAWTC 10 5.01
ATWCWT 7 5.01
ATTAYW 8 5.01
ATAYRG 7 5.01
ATARAC 5 5.01
ARTAWT 10 5.01
ACRTAS 5 5.01
TTSATW 7 5.00
RATGAW 8 5.00
CYSCTA 11 5.00
CWATAS 7 5.00
CTATYR 6 5.00
CGTWGW 6 5.00
ATAWGR 8 5.00
TGRTAR 7 4.99
TAYAGW 7 4.99
CTATTS 5 4.99
CTACWA 5 4.99
AYWCGA 5 4.99
ARTAYG 6 4.99
ACCTAW 4 4.99
TRTGGW 9 4.98
TAWTAY 8 4.98
TAGTSW 7 4.98
GAWTAR 8 4.98
CWCTAW 8 4.98
AYAGTS 8 4.98
ATATRG 5 4.98
ATAAYS 7 4.98
ATAATS 5 4.98
TAATCR 4 4.97
GTRTAS 7 4.97
CWCSTA 8 4.97
AWTAGR 8 4.97
AWRATC 9 4.97
AGAATA 4 4.97
TWCCWA 8 4.96
TTATSR 7 4.96
TRTGGY 10 4.96
TAGTAR 4 4.96
GATARR 7 4.96
CTATSY 6 4.96
ATYAYA 7 4.96
ATWCAW 6 4.96
ATCWTW 7 4.96
ATARRA 9 4.96
ATAGST 4 4.96
TAWGGW 7 4.95
TATCRY 5 4.95
AYYGTA 5 4.95
ARTARG 9 4.95
AGTASG 5 4.95
TTYATS 9 4.94
SATGAW 8 4.94
GGWSTA 9 4.94
GAYTAW 7 4.94
GATTAW 5 4.94
CTASSA 8 4.94
CTAGGW 6 4.94
CCRTAY 6 4.94
ATCWYA 7 4.94
ASACSA 9 4.94
TAYGGY 6 4.93
TASYAA 6 4.93
TACRAY 5 4.93
AYTATG 4 4.93
AWATGR 10 4.93
ATWYCA 8 4.93
ATCWWA 6 4.93
TTYATR 9 4.92
TAGTCA 3 4.92
CGWAGR 8 4.92
CATARY 6 4.92
AYATAR 7 4.92
ATYCTA 4 4.92
TTSGWA 7 4.91
TRCCWA 7 4.91
RTCTAS 7 4.91
AWGATW 7 4.91
ATRGYA 6 4.91
ATCRWT 5 4.91
TATWGY 7 4.90
CAAYAY 8 4.90
AYCACA 6 4.90
AWTAGS 8 4.90
ATAYCA 4 4.90
ATACCW 4 4.90
ACGTWG 4 4.90
TAATAR 5 4.89
GTAGGA 4 4.89
CRCTAY 7 4.89
CGWARA 8 4.89
CGWAGA 5 4.89
TAGGYA 4 4.88
TAACGW 3 4.88
GWACTA 4 4.88
CWATAC 4 4.88
CSATWA 7 4.88
ACTRTR 7 4.88
TAGGRW 8 4.87
RATATS 7 4.87
GWAGTA 5 4.87
AYACYA 7 4.87
AWCAYA 7 4.87
ATGRRT 7 4.87
TWGTAW 6 4.86
STATAS 10 4.86
GWWGTA 7 4.86
GTCTAS 5 4.86
ATCARW 7 4.86
AAYRAT 7 4.86
CATRAW 8 4.85
ATWWGG 9 4.85
ACATAR 4 4.85
TAGRTA 3 4.84
TAGGAW 5 4.84
RATTAY 7 4.84
CGWTGW 6 4.84
ATCYWA 6 4.84
YCATAR 7 4.83
GTARWA 8 4.83
CGAWGA 5 4.83
AYASGA 8 4.83
ATASCW 6 4.83
CTAATA 3 4.82
AWGAAW 10 4.82
ASCRTA 5 4.82
AGWWTA 8 4.82
TTTCAY 7 4.81
TACWCS 8 4.81
GGTRTA 5 4.81
CTATYW 7 4.81
CSTARG 9 4.81
AWCYTA 6 4.81
ATGYWT 7 4.81
ATCAWA 4 4.81
GATTWW 9 4.80
CTSYTA 10 4.80
ATWATS 8 4.80
ATAYGS 6 4.80
ATAGTA 2 4.80
AAYWAT 8 4.80
CTATAY 4 4.79
CCTAGR 8 4.79
AWTATS 7 4.79
ATTGAY 4 4.79
TARTCW 7 4.78
GAWGAA 8 4.78
AYAYGA 6 4.78
ATRSCA 7 4.78
ATARSA 7 4.78
ATARGR 8 4.78
AGWATA 5 4.78
TAGYAS 7 4.77
GAWTAY 7 4.77
GYAGTA 5 4.76
GTATWR 6 4.76
GATGAW 5 4.76
CAWCAC 6 4.76
AYATWG 7 4.76
ATRTGG 6 4.76
ATRATA 4 4.76
WAATCW 9 4.75
TTCWTY 12 4.75
TCCWAW 9 4.75
CYTAGR 10 4.75
CCTAGS 8 4.75
AWCART 7 4.75
ATTAWY 8 4.75
ASGTTR 7 4.75
ACWACR 7 4.75
TGRTAS 6 4.74
CGTASW 5 4.74
CCTARS 10 4.74
ATYGTA 3 4.74
ATWTCR 7 4.74
ATWGTG 5 4.74
CRTAGR 6 4.73
AWTATW 10 4.73
TWGGTW 6 4.72
TRATAY 6 4.72
TAGTAY 3 4.72
GTYCTW 9 4.72
AWTGAW 7 4.72
AWGWTA 6 4.72
TTATSS 7 4.71
TRTGAY 7 4.71
TGATAR 4 4.71
TCYTAW 8 4.71
TATTWR 8 4.71
GYASTA 7 4.71
GWCYTA 7 4.71
GTAWTW 7 4.71
CTSCTA 7 4.71
CGTAGR 4 4.71
CATAYY 6 4.71
AYAGGW 9 4.71
ATYART 7 4.71
ATWAYC 7 4.71
ATTCRW 5 4.71
ASTAYG 5 4.71
GTASYA 6 4.70
AATRTR 8 4.70
CYTAGS 10 4.69
CTWCGR 7 4.69
AYYACA 8 4.69
ATGCRT 4 4.69
ATGARW 8 4.69
TCCTAR 6 4.68
TAGTWS 6 4.68
GWCCWA 8 4.68
CTAYWA 7 4.68
AWTAYG 6 4.68
ATASYG 6 4.68
TTGATR 4 4.67
TAYTCS 8 4.67
TAGYGA 4 4.67
TAATAS 4 4.67
RTATAS 6 4.67
CTARSA 8 4.67
AYRATC 6 4.67
ATYATA 4 4.67
ATRSGA 7 4.67
ATGAAA 4 4.67
ACCAWA 5 4.67
CTAYCR 6 4.66
ATYGRT 5 4.66
ATRGRT 6 4.66
ACYTAY 6 4.66
ACWAYG 6 4.66
TATWCW 7 4.65
GYWGTA 8 4.65
CCYATY 10 4.65
AYWGTA 6 4.65
ATYGWT 5 4.65
ATYCAW 6 4.65
ATGYAY 6 4.65
ATCCWA 4 4.65
ACSTAT 3 4.65
ACAACR 5 4.65
GWAYTA 6 4.64
CTAYAS 8 4.64
CATCRY 7 4.64
TATTAR 5 5.02
CRTAWA 7 5.02
AWAATY 11 5.02
ATWTYA 9 5.02
AGTART 5 5.02
AWAWTC 10 5.01
ATWCWT 7 5.01
ATTAYW 8 5.01
ATAYRG 7 5.01
ATARAC 5 5.01
ARTAWT 10 5.01
ACRTAS 5 5.01
TTSATW 7 5.00
RATGAW 8 5.00
CYSCTA 11 5.00
CWATAS 7 5.00
CTATYR 6 5.00
CGTWGW 6 5.00
ATAWGR 8 5.00
TGRTAR 7 4.99
TAYAGW 7 4.99
CTATTS 5 4.99
CTACWA 5 4.99
AYWCGA 5 4.99
ARTAYG 6 4.99
ACCTAW 4 4.99
TRTGGW 9 4.98
TAWTAY 8 4.98
TAGTSW 7 4.98
GAWTAR 8 4.98
CWCTAW 8 4.98
AYAGTS 8 4.98
ATATRG 5 4.98
ATAAYS 7 4.98
ATAATS 5 4.98
TAATCR 4 4.97
GTRTAS 7 4.97
CWCSTA 8 4.97
AWTAGR 8 4.97
AWRATC 9 4.97
AGAATA 4 4.97
TWCCWA 8 4.96
TTATSR 7 4.96
TRTGGY 10 4.96
TAGTAR 4 4.96
GATARR 7 4.96
CTATSY 6 4.96
ATYAYA 7 4.96
ATWCAW 6 4.96
ATCWTW 7 4.96
ATARRA 9 4.96
ATAGST 4 4.96
TAWGGW 7 4.95
TATCRY 5 4.95
AYYGTA 5 4.95
ARTARG 9 4.95
AGTASG 5 4.95
TTYATS 9 4.94
SATGAW 8 4.94
GGWSTA 9 4.94
GAYTAW 7 4.94
GATTAW 5 4.94
CTASSA 8 4.94
CTAGGW 6 4.94
CCRTAY 6 4.94
ATCWYA 7 4.94
ASACSA 9 4.94
TAYGGY 6 4.93
TASYAA 6 4.93
TACRAY 5 4.93
AYTATG 4 4.93
AWATGR 10 4.93
ATWYCA 8 4.93
ATCWWA 6 4.93
TTYATR 9 4.92
TAGTCA 3 4.92
CGWAGR 8 4.92
CATARY 6 4.92
AYATAR 7 4.92
ATYCTA 4 4.92
TTSGWA 7 4.91
TRCCWA 7 4.91
RTCTAS 7 4.91
AWGATW 7 4.91
ATRGYA 6 4.91
ATCRWT 5 4.91
TATWGY 7 4.90
CAAYAY 8 4.90
AYCACA 6 4.90
AWTAGS 8 4.90
ATAYCA 4 4.90
ATACCW 4 4.90
ACGTWG 4 4.90
TAATAR 5 4.89
GTAGGA 4 4.89
CRCTAY 7 4.89
CGWARA 8 4.89
CGWAGA 5 4.89
TAGGYA 4 4.88
TAACGW 3 4.88
GWACTA 4 4.88
CWATAC 4 4.88
CSATWA 7 4.88
ACTRTR 7 4.88
TAGGRW 8 4.87
RATATS 7 4.87
GWAGTA 5 4.87
AYACYA 7 4.87
AWCAYA 7 4.87
ATGRRT 7 4.87
TWGTAW 6 4.86
STATAS 10 4.86
GWWGTA 7 4.86
GTCTAS 5 4.86
ATCARW 7 4.86
AAYRAT 7 4.86
CATRAW 8 4.85
ATWWGG 9 4.85
ACATAR 4 4.85
TAGRTA 3 4.84
TAGGAW 5 4.84
RATTAY 7 4.84
CGWTGW 6 4.84
ATCYWA 6 4.84
YCATAR 7 4.83
GTARWA 8 4.83
CGAWGA 5 4.83
AYASGA 8 4.83
ATASCW 6 4.83
CTAATA 3 4.82
AWGAAW 10 4.82
ASCRTA 5 4.82
AGWWTA 8 4.82
TTTCAY 7 4.81
TACWCS 8 4.81
GGTRTA 5 4.81
CTATYW 7 4.81
CSTARG 9 4.81
AWCYTA 6 4.81
ATGYWT 7 4.81
ATCAWA 4 4.81
GATTWW 9 4.80
CTSYTA 10 4.80
ATWATS 8 4.80
ATAYGS 6 4.80
ATAGTA 2 4.80
AAYWAT 8 4.80
CTATAY 4 4.79
CCTAGR 8 4.79
AWTATS 7 4.79
ATTGAY 4 4.79
TARTCW 7 4.78
GAWGAA 8 4.78
AYAYGA 6 4.78
ATRSCA 7 4.78
ATARSA 7 4.78
ATARGR 8 4.78
AGWATA 5 4.78
TAGYAS 7 4.77
GAWTAY 7 4.77
GYAGTA 5 4.76
GTATWR 6 4.76
GATGAW 5 4.76
CAWCAC 6 4.76
AYATWG 7 4.76
ATRTGG 6 4.76
ATRATA 4 4.76
WAATCW 9 4.75
TTCWTY 12 4.75
TCCWAW 9 4.75
CYTAGR 10 4.75
CCTAGS 8 4.75
AWCART 7 4.75
ATTAWY 8 4.75
ASGTTR 7 4.75
ACWACR 7 4.75
TGRTAS 6 4.74
CGTASW 5 4.74
CCTARS 10 4.74
ATYGTA 3 4.74
ATWTCR 7 4.74
ATWGTG 5 4.74
CRTAGR 6 4.73
AWTATW 10 4.73
TWGGTW 6 4.72
TRATAY 6 4.72
TAGTAY 3 4.72
GTYCTW 9 4.72
AWTGAW 7 4.72
AWGWTA 6 4.72
TTATSS 7 4.71
TRTGAY 7 4.71
TGATAR 4 4.71
TCYTAW 8 4.71
TATTWR 8 4.71
GYASTA 7 4.71
GWCYTA 7 4.71
GTAWTW 7 4.71
CTSCTA 7 4.71
CGTAGR 4 4.71
CATAYY 6 4.71
AYAGGW 9 4.71
ATYART 7 4.71
ATWAYC 7 4.71
ATTCRW 5 4.71
ASTAYG 5 4.71
GTASYA 6 4.70
AATRTR 8 4.70
CYTAGS 10 4.69
CTWCGR 7 4.69
AYYACA 8 4.69
ATGCRT 4 4.69
ATGARW 8 4.69
TCCTAR 6 4.68
TAGTWS 6 4.68
GWCCWA 8 4.68
CTAYWA 7 4.68
AWTAYG 6 4.68
ATASYG 6 4.68
TTGATR 4 4.67
TAYTCS 8 4.67
TAGYGA 4 4.67
TAATAS 4 4.67
RTATAS 6 4.67
CTARSA 8 4.67
AYRATC 6 4.67
ATYATA 4 4.67
ATRSGA 7 4.67
ATGAAA 4 4.67
ACCAWA 5 4.67
CTAYCR 6 4.66
ATYGRT 5 4.66
ATRGRT 6 4.66
ACYTAY 6 4.66
ACWAYG 6 4.66
TATWCW 7 4.65
GYWGTA 8 4.65
CCYATY 10 4.65
AYWGTA 6 4.65
ATYGWT 5 4.65
ATYCAW 6 4.65
ATGYAY 6 4.65
ATCCWA 4 4.65
ACSTAT 3 4.65
ACAACR 5 4.65
GWAYTA 6 4.64
CTAYAS 8 4.64
CATCRY 7 4.64
References
[1] Heinemeyer, T., Chen, X., Karas, H., Kel, A. E., Kel, O. V., Liebich, I., Meinhardt, T., Reuter, I., Schacherer, F., Wingender, E. Expanding the TRANSFAC database towards an expert system of regulatory molecular mechanisms Nucleic Acids Res. 27, 318-322, (1999). PubMed: 99063727
[2] Timothy L. Bailey and Charles Elkan, "Fitting a mixture model by expectation maximization to discover motifs in biopolymers", Proceedings of the Second International Conference on Intelligent Systems for Molecular Biology, pp. 28-36, AAAI Press, Menlo Park, California, 1994. [postscript] [pdf]
[3] The Sequence Manipulation. (2010). The Gene Infinity. Retrieved March 2010, from DNA Restriction Enzyme Summary: http://www.geneinfinity.org/sms_remap.html
[4] Stothard P (2000) The Sequence Manipulation Suite: JavaScript programs for analyzing and formatting protein and DNA sequences. Biotechniques 28:1102-1104.
[5] Xiaoli Dong, Paul Stothard, Ian J. Forsythe, and David S. Wishart "PlasMapper: a web server for drawing and auto-annotating plasmid maps" Nucleic Acids Res. 2004 Jul 1;32(Web Server issue):W660-4.
[2] Timothy L. Bailey and Charles Elkan, "Fitting a mixture model by expectation maximization to discover motifs in biopolymers", Proceedings of the Second International Conference on Intelligent Systems for Molecular Biology, pp. 28-36, AAAI Press, Menlo Park, California, 1994. [postscript] [pdf]
[3] The Sequence Manipulation. (2010). The Gene Infinity. Retrieved March 2010, from DNA Restriction Enzyme Summary: http://www.geneinfinity.org/sms_remap.html
[4] Stothard P (2000) The Sequence Manipulation Suite: JavaScript programs for analyzing and formatting protein and DNA sequences. Biotechniques 28:1102-1104.
[5] Xiaoli Dong, Paul Stothard, Ian J. Forsythe, and David S. Wishart "PlasMapper: a web server for drawing and auto-annotating plasmid maps" Nucleic Acids Res. 2004 Jul 1;32(Web Server issue):W660-4.