Subacute Necrotizing Encephalomyelopathy and MT-CO1
Protein Homology
The amino acid sequence of Cytochrome c Oxidase Subunit 1(human) was blasted to find the the closest 17 orthologs [5]. The orthologs were then collected and aligned using various free online alignment programs. This analysis was then compiled into phylogenetic trees.
Each alignment program yielded similar results, however based on the algorithms, values of homology varied slightly.
The FASTA amino acid sequences of all orthologs are compiled in the text file below.
Each alignment program yielded similar results, however based on the algorithms, values of homology varied slightly.
The FASTA amino acid sequences of all orthologs are compiled in the text file below.
| protein_orthologs.txt |
The species names of the protein orthologs are used on the alignment. The common name is matched below, along with the Entrez Protein ID in (parenthesis).
Species
Arabadopsis thaliana
Oryza sativa
Caenorhabditis elegans
Drosophila melanogaster
Anopheles gambiae
Gallus gallus
Danio rerio
Canis lupus familiaris
Homo sapiens
Pan troglodytes
Bos taurus
Mus musculus
Rattus norvegicus
Saccharomyces cerevisiae
Kluyveromyces lactis
Ashbya gossypii
Schizosaccharomyces pombe
Plasmodium falciparum
Arabadopsis thaliana
Oryza sativa
Caenorhabditis elegans
Drosophila melanogaster
Anopheles gambiae
Gallus gallus
Danio rerio
Canis lupus familiaris
Homo sapiens
Pan troglodytes
Bos taurus
Mus musculus
Rattus norvegicus
Saccharomyces cerevisiae
Kluyveromyces lactis
Ashbya gossypii
Schizosaccharomyces pombe
Plasmodium falciparum
Common Name (Entrez Protein ID)
Arabidopsis (NP_085587.1)
Asian Rice (YP_514675.1)
Nematode (NP_006961.1)
Fruit Fly (NP_008278.1)
Mosquitoe (NP_008070.1)
Red Junglefowl (NP_006917.1)
Zebrafish (NP_059333.1)
Domesticated Dog (NP_008473.4)
Human (YP_003024028)
Common/Robust Chimpanzee (NP_008188.1)
Domesticated Cattle (YP_209207.1)
House Mouse (NP_904330.1)
Norway Rat (YP_665631.1)
Budding Yeast (NP_009305.1)
Kluyveromyces Yeast (YP_054500.1)
Filamentous Fungus (NP_987079.1)
Fission Yeast (NP_039499.1)
Parasite (NP_059667.1)
Arabidopsis (NP_085587.1)
Asian Rice (YP_514675.1)
Nematode (NP_006961.1)
Fruit Fly (NP_008278.1)
Mosquitoe (NP_008070.1)
Red Junglefowl (NP_006917.1)
Zebrafish (NP_059333.1)
Domesticated Dog (NP_008473.4)
Human (YP_003024028)
Common/Robust Chimpanzee (NP_008188.1)
Domesticated Cattle (YP_209207.1)
House Mouse (NP_904330.1)
Norway Rat (YP_665631.1)
Budding Yeast (NP_009305.1)
Kluyveromyces Yeast (YP_054500.1)
Filamentous Fungus (NP_987079.1)
Fission Yeast (NP_039499.1)
Parasite (NP_059667.1)
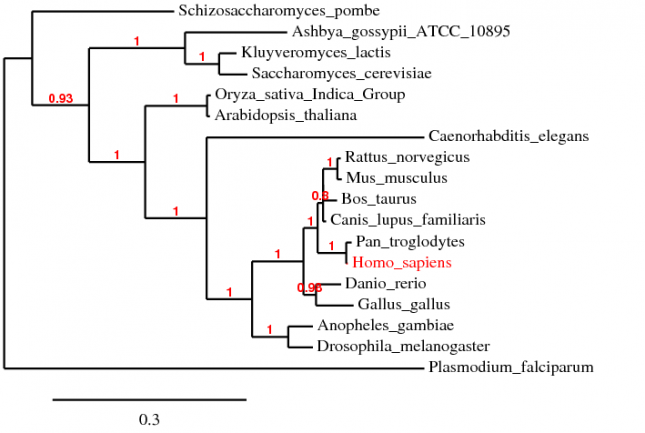
Alignment using ClustalW
The alignment using ClustalW 2.0.3 is compiled into the text file below [1]. The default settings were used during the alignmet. The phylogenetic tree is shown below.
| mt-co1_clustalw_alignment.txt |
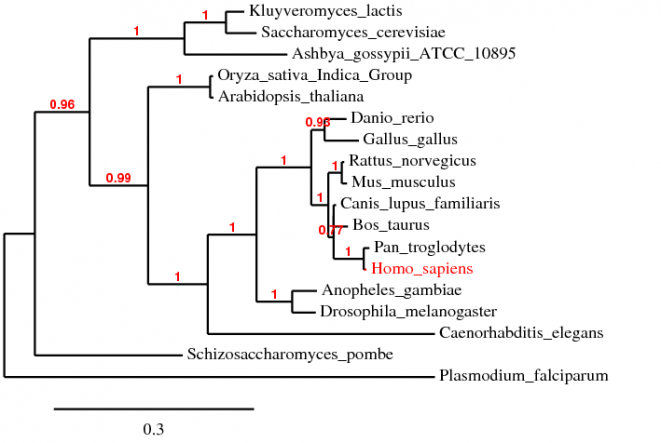
Alignment using MUSCLE
The alignment using MUSCLE 3.7 is compiled into the text file below [2]. The default settings were used during the alignmet. The phylogenetic tree is shown below.
| mt-co1_muscle_alignment.txt |
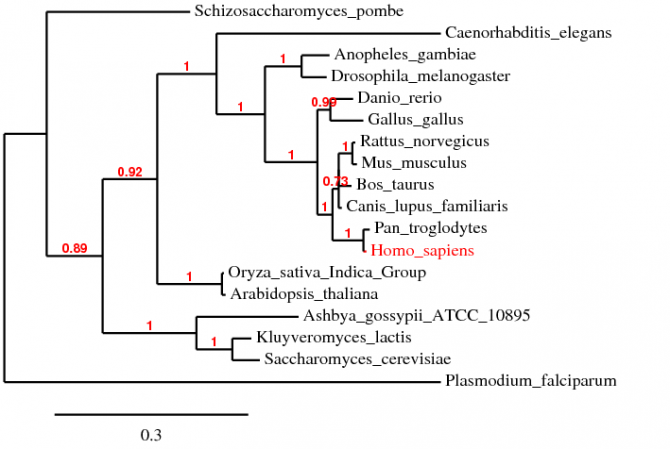
Alignment using T-Coffee
The alignment using T-Coffe 6.85 is compiled into the text file below [3]. The default settings were used during the alignmet. The phylogenetic tree is shown below.
| mt-co1_t-coffee_alignment.txt |
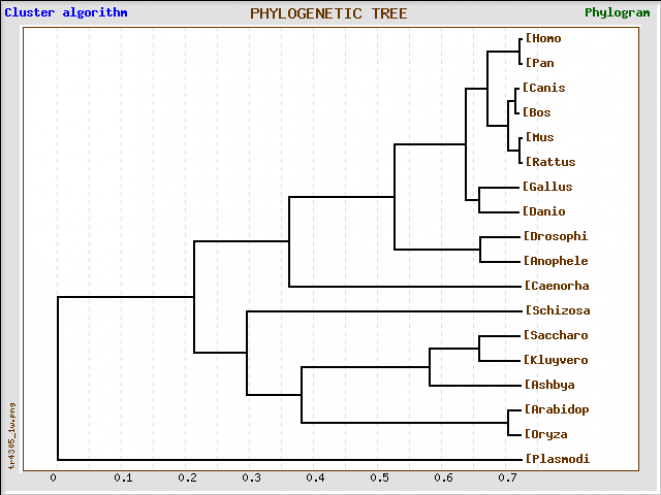
Alignment using GeneBee
The alignment using GeneBee is compiled into the word file below [4]. The default settings were used during the alignmet. The phylogenetic tree is shown below.
| mt-co1_genebee_alignment.docx |
Analysis using STRING
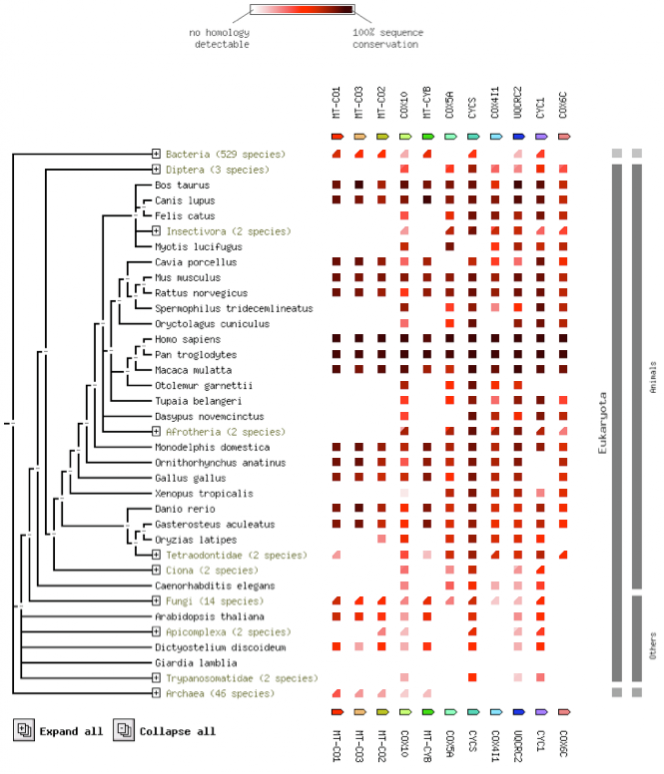
The alignment using STRING is compiled in the image below [6]. Using the amino acid sequence of Cytochrome c Oxidase Subunit 1, STRING generated a large list of orthologs, with the species shown in a phylognic tree on the left. Across the horizontal of the image are a series of protein sequences that are known to interact with Cytochrome c Oxidase Subunit 1. Those amino acid sequences were also compared and colored based on how conserved the respective sequence is (Darker = More conserved).
References
[1] GForge Project. Methodes et Algorithms pour la Bio-Informatique. Retrieved March 23, 2010, from ClustalW 2.0.3: http://www.phylogeny.fr/version2_cgi/one_task.cgi?task_type=clustalw
[2] GForge Project. Methodes et Algorithms pour la Bio-Informatique. Retrieved March 23, 2010, from MUSCLE 3.7: http://www.phylogeny.fr/version2_cgi/one_task.cgi?task_type=muscle
[3] GForge Project. Methodes et Algorithms pour la Bio-Informatique. Retrieved March 23, 2010, from T-Coffee 6.85: http://www.phylogeny.fr/version2_cgi/one_task.cgi?task_type=tcoffee
[4] GeneBee Group. GeneBee - Molecular Biology Server. Retrieved March 23, 2010, from Alibee - Multiple Alignment: http://www.genebee.msu.su/services/malign_reduced.html
[5] National Center for Biotechnology Information. Basic Local Assignment Search Tool. Retrieved March 23, 2010, from Blastp: http://blast.ncbi.nlm.nih.gov/Blast.cgi?PROGRAM=blastp&BLAST_PROGRAMS=blastp&PAGE_TYPE=BlastSearch&SHOW_DEFAULTS=on&LINK_LOC=blasthome
[6] STRING 8.2. Search Tool for the Retrieval of Interacting Genes/Proteins. Retrieved March 24, 2010, from STRING: http://string.embl.de/newstring_cgi/show_input_page.pl?UserId=Ud2Uew0uKlBW&sessionId=vsE5HOz75lGb&info_box_type_input_page=general
References specific to phylogenetic trees (other than Genebee, and STRING):
1. Dereeper A.*, Guignon V.*, Blanc G., Audic S., Buffet S., Chevenet F., Dufayard J.F., Guindon S., Lefort V., Lescot M., Claverie J.M., Gascuel O. Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 2008 Jul 1;36(Web Server issue):W465-9. Epub 2008 Apr 19. (PubMed) *: joint first authors
2. Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, Mar 19;32(5):1792-7. (PubMed)
3. Castresana J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol. 2000, Apr;17(4):540-52. (PubMed)
4. Huelsenbeck JP., Ronquist F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics. 2001, Aug;17(8):754-5. (PubMed)
5. Chevenet F., Brun C., Banuls AL., Jacq B., Chisten R. TreeDyn: towards dynamic graphics and annotations for analyses of trees. BMC Bioinformatics. 2006, Oct 10;7:439. (PubMed)
[2] GForge Project. Methodes et Algorithms pour la Bio-Informatique. Retrieved March 23, 2010, from MUSCLE 3.7: http://www.phylogeny.fr/version2_cgi/one_task.cgi?task_type=muscle
[3] GForge Project. Methodes et Algorithms pour la Bio-Informatique. Retrieved March 23, 2010, from T-Coffee 6.85: http://www.phylogeny.fr/version2_cgi/one_task.cgi?task_type=tcoffee
[4] GeneBee Group. GeneBee - Molecular Biology Server. Retrieved March 23, 2010, from Alibee - Multiple Alignment: http://www.genebee.msu.su/services/malign_reduced.html
[5] National Center for Biotechnology Information. Basic Local Assignment Search Tool. Retrieved March 23, 2010, from Blastp: http://blast.ncbi.nlm.nih.gov/Blast.cgi?PROGRAM=blastp&BLAST_PROGRAMS=blastp&PAGE_TYPE=BlastSearch&SHOW_DEFAULTS=on&LINK_LOC=blasthome
[6] STRING 8.2. Search Tool for the Retrieval of Interacting Genes/Proteins. Retrieved March 24, 2010, from STRING: http://string.embl.de/newstring_cgi/show_input_page.pl?UserId=Ud2Uew0uKlBW&sessionId=vsE5HOz75lGb&info_box_type_input_page=general
References specific to phylogenetic trees (other than Genebee, and STRING):
1. Dereeper A.*, Guignon V.*, Blanc G., Audic S., Buffet S., Chevenet F., Dufayard J.F., Guindon S., Lefort V., Lescot M., Claverie J.M., Gascuel O. Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 2008 Jul 1;36(Web Server issue):W465-9. Epub 2008 Apr 19. (PubMed) *: joint first authors
2. Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, Mar 19;32(5):1792-7. (PubMed)
3. Castresana J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol. 2000, Apr;17(4):540-52. (PubMed)
4. Huelsenbeck JP., Ronquist F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics. 2001, Aug;17(8):754-5. (PubMed)
5. Chevenet F., Brun C., Banuls AL., Jacq B., Chisten R. TreeDyn: towards dynamic graphics and annotations for analyses of trees. BMC Bioinformatics. 2006, Oct 10;7:439. (PubMed)