Subacute Necrotizing Encephalomyelopathy and MT-CO1
Cytochrome c Oxidase Subunit 1 Domains
The amino acid sequence of Cytochrome c Oxidase subunit 1 (file below) was analyzed based on domains via the free online biotechnology engines below.
| cytochrome_c_oxidase_subunit_i_protein_sequence.docx |
Codon Bias
The codon bias is said to be important in the comparison of orthologs, specifically inserting new genes into a model organism. The codon bias for the MT-CO1 geneis shown below [1,2]. All other orthologs of the gene can have their sequence analyzed based on the codon bias using Gene Infinity Codon Usage.
Amino Codon Number Percent
Acid
Ala GCG 1.00 0.195
GCA 13.00 2.529
GCT 11.00 2.140
GCC 15.00 2.918
Cys TGT 0.00 0.00
TGC 1.00 0.195
Asp GAT 2.00 0.389
GAC 13.00 2.529
Glu GAG 3.00 0.584
GAA 7.00 1.362
Phe TTT 12.00 2.335
TTC 29.00 5.642
Gly GGG 3.00 0.584
GGA 19.00 3.696
GGT 7.00 1.362
Gly GGC 17.00 3.307
His CAT 4.00 0.778
CAC 14.00 2.724
Ile ATA 25.00 4.864
ATT 12.00 2.335
ATC 26.00 5.058
Lys AAG 1.00 0.195
AAA 9.00 1.751
Leu TTG 0.00 0.00
TTA 7.00 1.362
CTG 4.00 0.778
CTA 33.00 6.420
CTT 5.00 0.973
CTC 13.00 2.529
Met ATG 7.00 1.362
Acid
Ala GCG 1.00 0.195
GCA 13.00 2.529
GCT 11.00 2.140
GCC 15.00 2.918
Cys TGT 0.00 0.00
TGC 1.00 0.195
Asp GAT 2.00 0.389
GAC 13.00 2.529
Glu GAG 3.00 0.584
GAA 7.00 1.362
Phe TTT 12.00 2.335
TTC 29.00 5.642
Gly GGG 3.00 0.584
GGA 19.00 3.696
GGT 7.00 1.362
Gly GGC 17.00 3.307
His CAT 4.00 0.778
CAC 14.00 2.724
Ile ATA 25.00 4.864
ATT 12.00 2.335
ATC 26.00 5.058
Lys AAG 1.00 0.195
AAA 9.00 1.751
Leu TTG 0.00 0.00
TTA 7.00 1.362
CTG 4.00 0.778
CTA 33.00 6.420
CTT 5.00 0.973
CTC 13.00 2.529
Met ATG 7.00 1.362
Amino Codon Number Percent
Acid
Asn AAT 4.00 0.778
AAC 13.00 2.529
Pro CCG 0.00 0.00
CCA 6.00 1.167
CCT 5.00 0.973
CCC 18.00 3.502
Gln CAG 1.00 0.195
CAA 5.00 0.973
Arg AGG 0.00 0.00
AGA 1.00 0.195
CGG 0.00 0.00
CGA 4.00 0.778
CGT 2.00 0.389
CGC 2.00 0.389
Ser AGT 0.00 0.00
AGC 4.00 0.778
TCG 2.00 0.389
TCA 7.00 1.362
TCT 9.00 1.751
TCC 10.00 1.946
Thr ACG 2.00 0.389
ACA 16.00 3.113
ACT 5.00 0.973
ACC 11.00 2.140
Val GTG 3.00 0.584
GTA 17.00 3.307
GTT 4.00 0.778
GTC 12.00 2.335
Trp TGG 0.00 0.00
Tyr TAT 4.00 0.778
TAC 18.00 3.502
End TGA 16.00 3.113
TAG 0.00 0.00
TAA 0.00 0.00
Acid
Asn AAT 4.00 0.778
AAC 13.00 2.529
Pro CCG 0.00 0.00
CCA 6.00 1.167
CCT 5.00 0.973
CCC 18.00 3.502
Gln CAG 1.00 0.195
CAA 5.00 0.973
Arg AGG 0.00 0.00
AGA 1.00 0.195
CGG 0.00 0.00
CGA 4.00 0.778
CGT 2.00 0.389
CGC 2.00 0.389
Ser AGT 0.00 0.00
AGC 4.00 0.778
TCG 2.00 0.389
TCA 7.00 1.362
TCT 9.00 1.751
TCC 10.00 1.946
Thr ACG 2.00 0.389
ACA 16.00 3.113
ACT 5.00 0.973
ACC 11.00 2.140
Val GTG 3.00 0.584
GTA 17.00 3.307
GTT 4.00 0.778
GTC 12.00 2.335
Trp TGG 0.00 0.00
Tyr TAT 4.00 0.778
TAC 18.00 3.502
End TGA 16.00 3.113
TAG 0.00 0.00
TAA 0.00 0.00
SMART Search
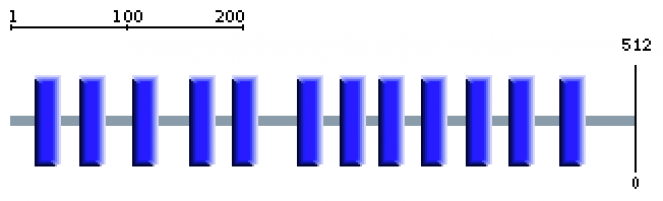
SMART was used to determine known domains found within the amino acid sequence of Cytochrome c Oxidase subunit 1. A total of 12 trans membrane domains were found (shown below) through the TMHMM Server v 2.0.
PROSITE Search
PROSITE was used to determine known domains found within the amino acid sequence of Cytochrome c Oxidase subunit 1. A Heme-copper oxidase catalytic subunit, copper B binding region signature domain (PS00077) was found from amino acids 236-291.
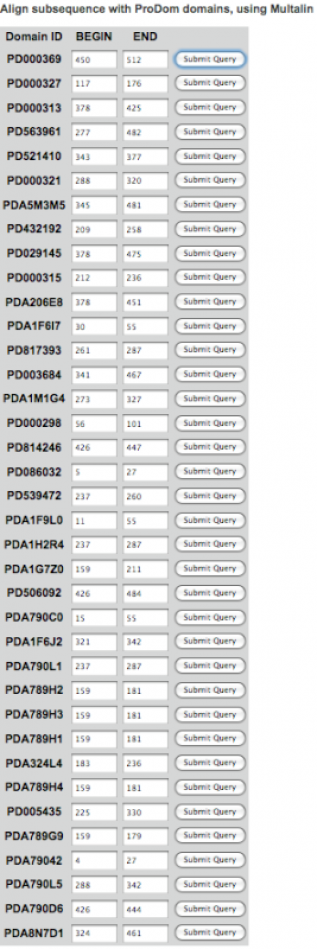
ProDom Search
ProDom was used to determine known domains found within the amino acid sequence of Cytochrome c Oxidase subunit 1. A large number of domains were found using 3 different algorithms (shown below). However, most "Submit Query" links were unusable, and did not yield further information of the domain. Those that did have active links were of little use, as they only aligned other sequences with the domain, rather than give the common function.
References
[1] Stothard P (2000) The Sequence Manipulation Suite: JavaScript programs for analyzing and formatting protein and DNA sequences. Biotechniques 28:1102-1104.
[2] The Sequence Manipulation. (2010). The Gene Infinity. Retrieved March 2010, from DNA Codon Usage:http://www.geneinfinity.org/sms_codonusage.html
[2] The Sequence Manipulation. (2010). The Gene Infinity. Retrieved March 2010, from DNA Codon Usage:http://www.geneinfinity.org/sms_codonusage.html